ResearchResearch OverviewTechnologyplexDIAApplicationsNeurodegenerative DiseasesConnect & LearnWorkshops & EventsPublicationsVideos
CareersBlogVisit UsContact
About Our Research
We aim to increase the throughput of mass spectrometry proteomics 100-1000x
We aim to analyze the proteomes of more than 30,000 single cells or small-input samples per day without sacrificing accuracy.
Technology
We have already demonstrated a proof-of-principle through the development of plexDIA. That endeavor increased the sensitivity of low-input samples using commercially available reagents. We now aim to scale the throughput by 100-1000 fold by synergistically optimizing mass spectrometry methods, creation of novel barcodes, and implementing deep learning analysis. This multiplicative increase in throughput arises from the simultaneous parallelization of both samples (single cells per unit time) and the depth of proteome coverage (peptides quantified per sample). This inspires the name of this FRO, Parallel Squared.
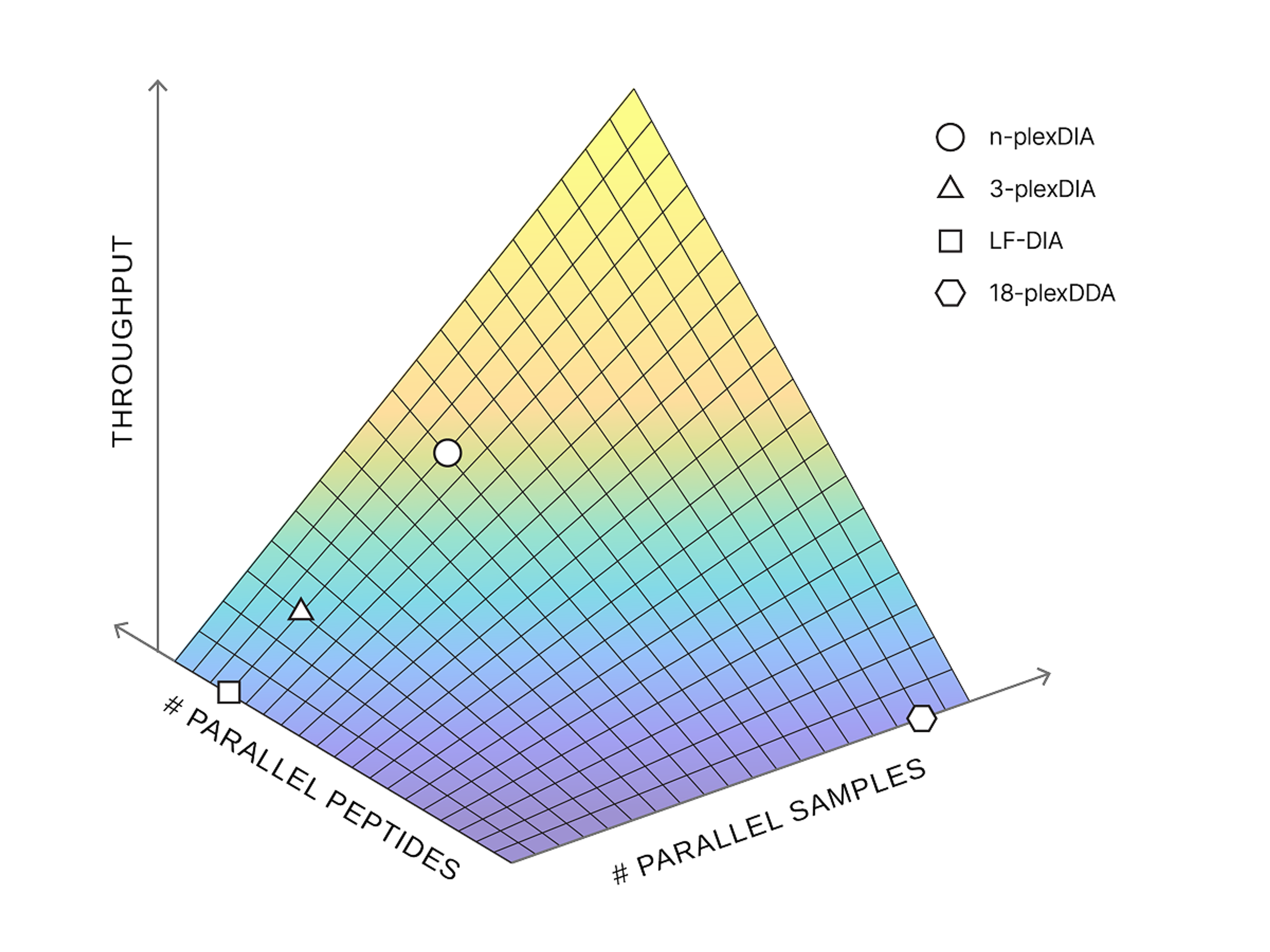
plexDIA
plexDIA is a multiplexing method that triples proteomics throughput from tiny samples without losing accuracy, enabling fast, high-coverage analysis—even at the single-cell level.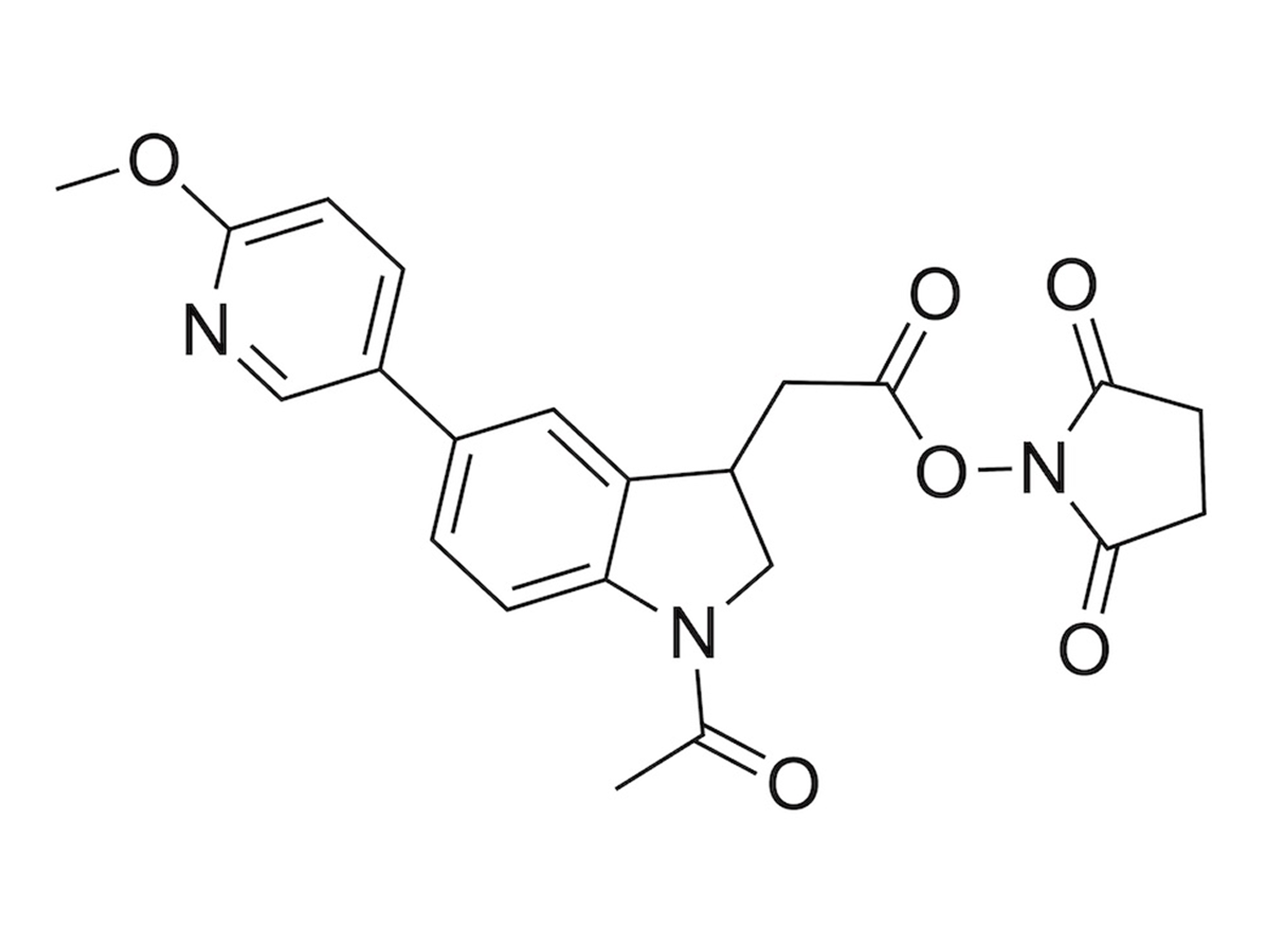
PSMtagComing Soon
A new mass tag enables 9-plexDIA with better sequencing and phosphorylation analysis, allowing high-throughput proteomics of over 1,000 samples per day.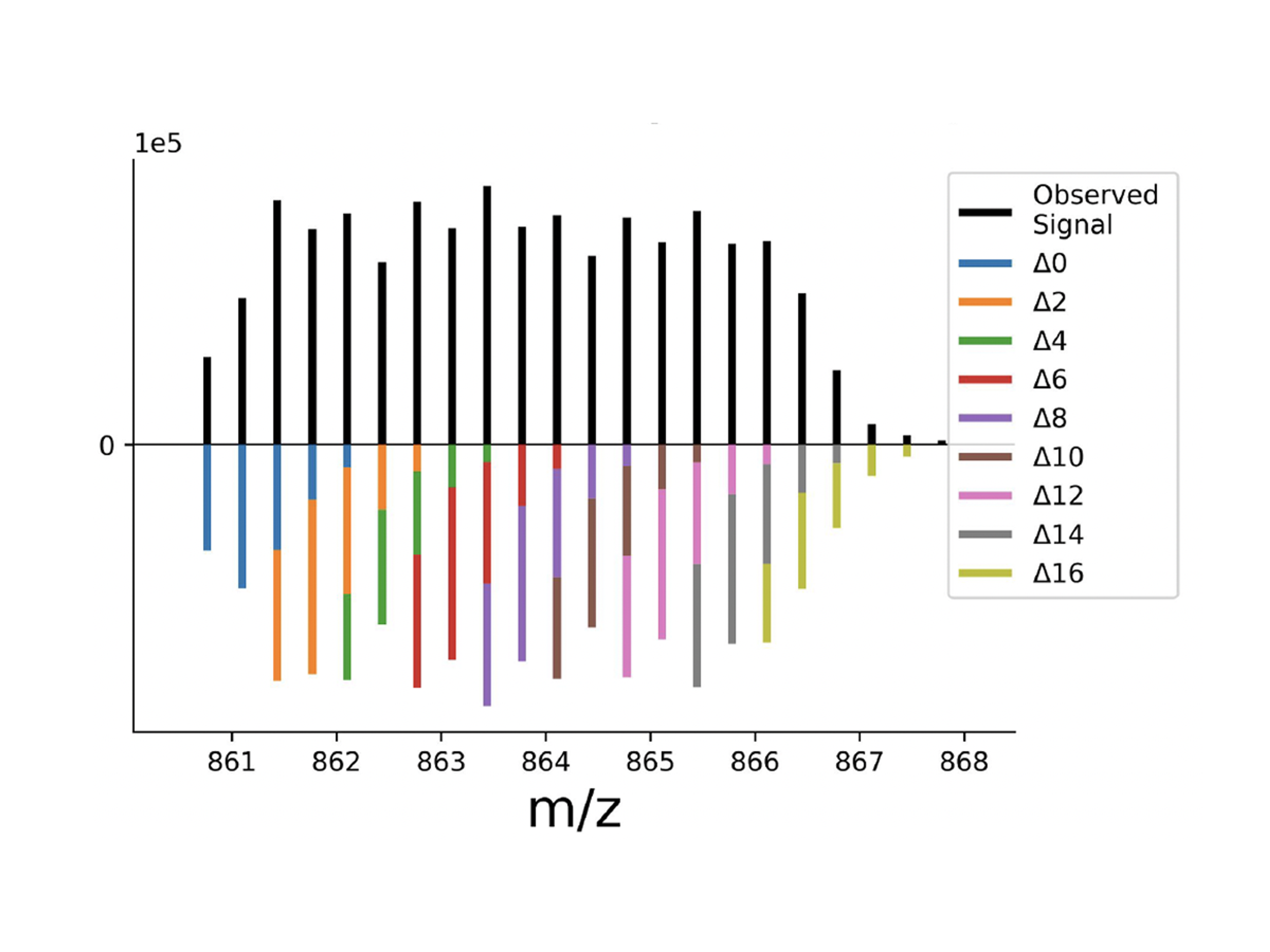
JModComing Soon
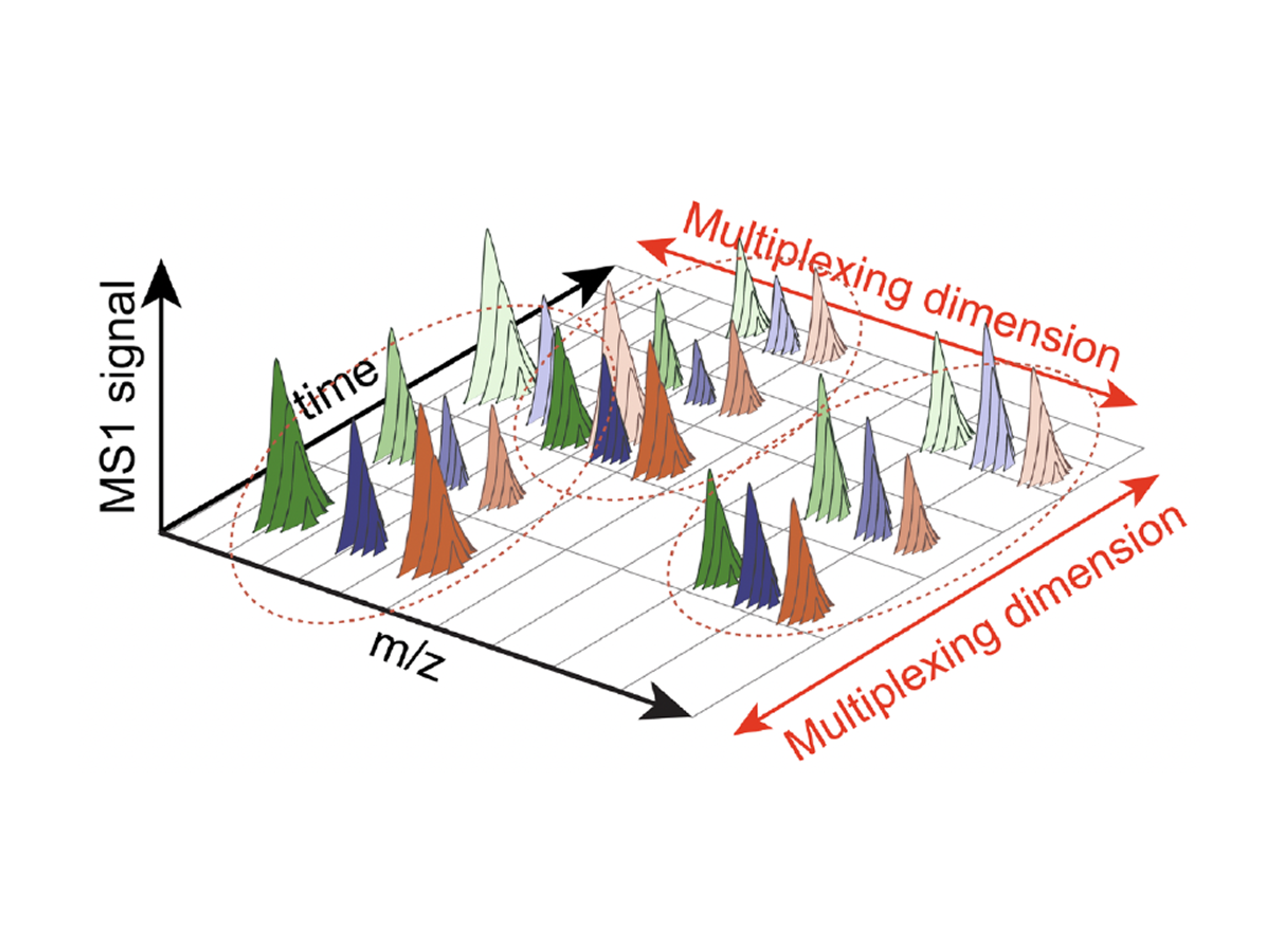
TimePlexComing Soon
Applications
Neurodegenerative Diseases
To better understand Alzheimer’s, our approach analyzes protein data at two scales—brain voxels to track spatial disease progression, and single cells to reveal how diverse cell types are differently affected.
Aging of Immune Cells
To uncover how aging affects immunity, we profile immune cells across diverse ages and backgrounds—using bulk analysis for deep proteomic insight and single-cell analysis to map individual cellular changes.A Convergent Research FRO
© 2025 Parallel Squared Technology Institute All rights reserved.