Initial recommendations for performing, benchmarking and reporting single-cell proteomics experiments
Laurent Gatto, Ruedi Aebersold, Juergen Cox, Vadim Demichev, Jason Derks, Edward Emmott, Alexander M. Franks, Alexander R. Ivanov, Ryan T. Kelly, Luke Khoury, Andrew Leduc, Michael J. MacCoss, Peter Nemes, David H. Perlman, Aleksandra A. Petelski, Christopher M. Rose, Erwin M. Schoof, Jennifer Van Eyk, Christophe Vanderaa, John R. Yates III & Nikolai Slavov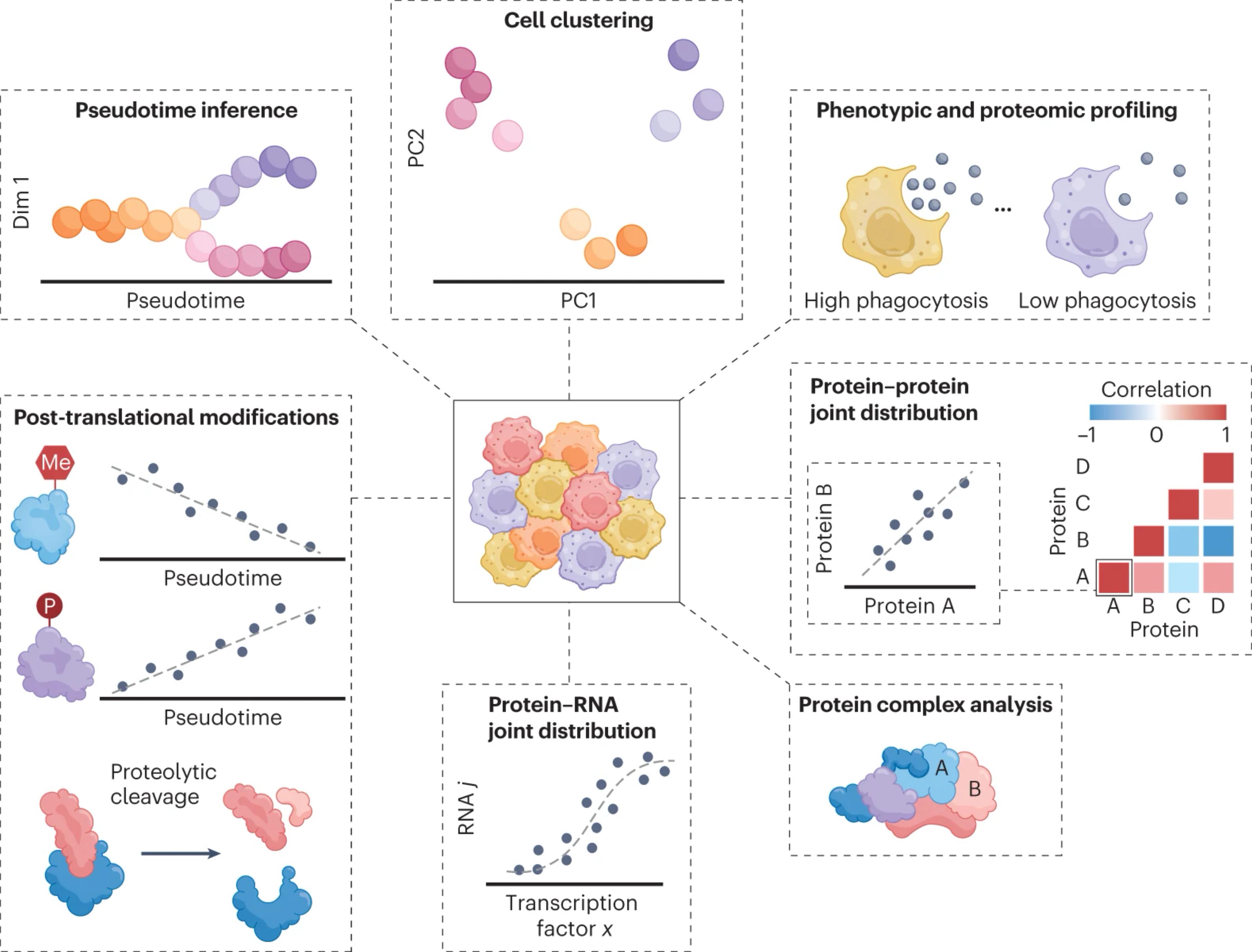
Abstract
Analyzing proteins from single cells by tandem mass spectrometry (MS) has recently become technically feasible. While such analysis has the potential to accurately quantify thousands of proteins across thousands of single cells, the accuracy and reproducibility of the results may be undermined by numerous factors affecting experimental design, sample preparation, data acquisition and data analysis. We expect that broadly accepted community guidelines and standardized metrics will enhance rigor, data quality and alignment between laboratories. Here we propose best practices, quality controls and data-reporting recommendations to assist in the broad adoption of reliable quantitative workflows for single-cell proteomics. Resources and discussion forums are available at https://single-cell.net/guidelines.