Barcodes and the future of measuring proteins
Increasing the rate at which we measure proteins increases the rate at which we can discover new biology and solve disease.
What does the future hold for how we study proteins?
What technology will we use? How easy will that technology be to use?
How expensive? How fast?
Here’s why these questions are important: Increasing the rate at which we measure proteins increases the rate at which we can discover new biology and solve disease.
You can try to predict the future based on current trends. Or you can try to alter the trends yourself. This conceit is the ambition of focused research organizations.
Parallel Squared Technology Institute (PTI) was founded as a focused research organization to buck trends by increasing the speed and decreasing the cost of protein technology 100-1000x in just five years.
How feasible is this ambition? A partial answer to that question lies in our ability to create 100s of barcodes.
These are not grocery-store barcodes, though they serve the same purpose: to uniquely identify. Instead of marking collections of produce, our barcodes uniquely identify collections of proteins (often called 'proteomes'). Proteomes can come from a huge range of biological sources: from a blood draw of an athlete to a single neuron from an Alzheimer's disease patient.
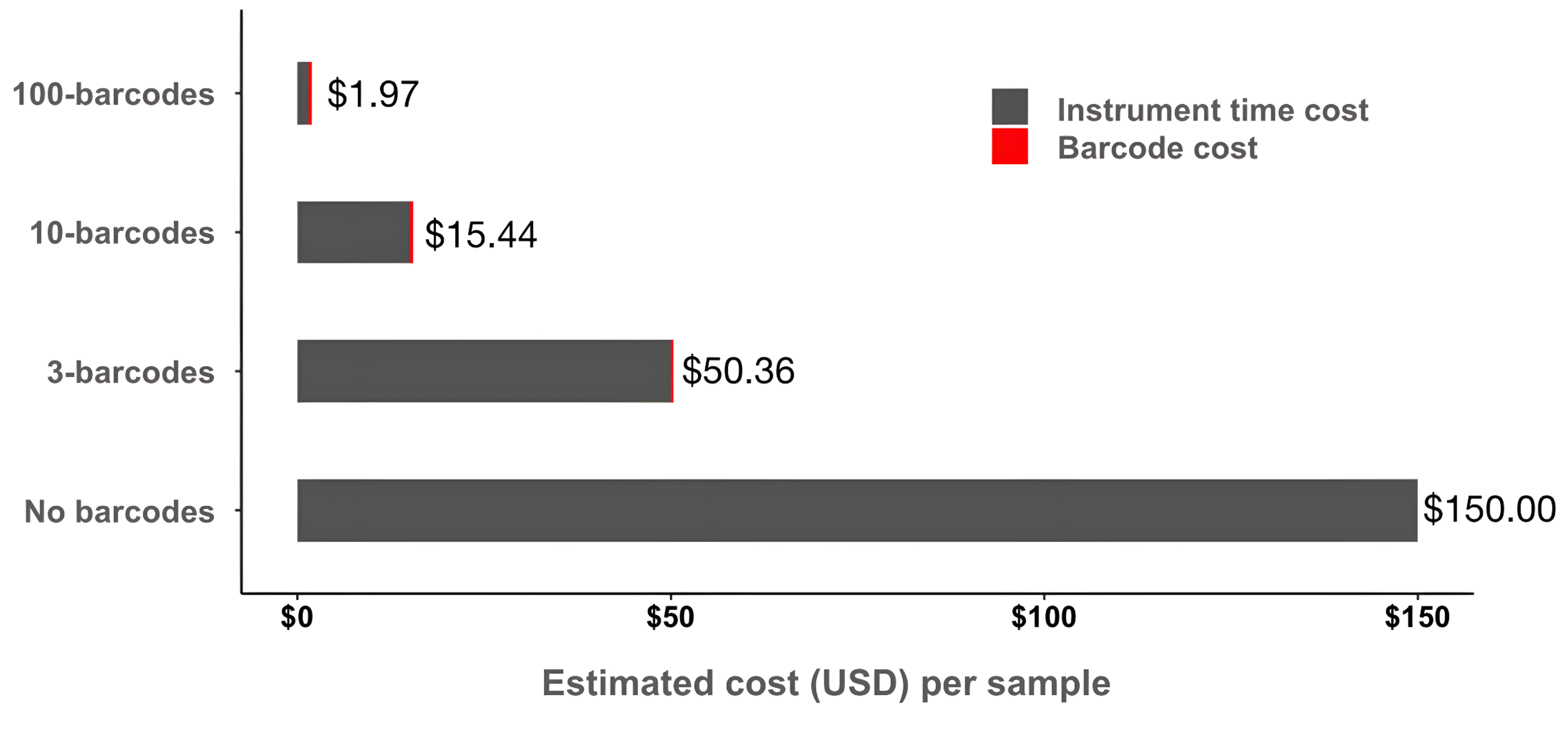
By applying barcodes, hundreds or even thousands of proteomes can be simultaneously analyzed, increasing speed and decreasing cost almost proportional to the number of barcodes.
In 'How to design 1000-plex mass tags using the differential mass defect', we explore how many barcodes different molecules can make. We find barcodes in the hundreds, an order of magnitude greater than state-of-the-art, are achievable using molecules comparable in size to existing commercial tags, and that going beyond hundreds may require larger molecules.
We are convinced that we are standing in an orchard of low-hanging fruit for creating 100s of barcodes. Creating 1000s of barcodes will be more challenging, but clearly possible as well.
Along the way we describe a new term we found helpful and even how the most famous equation of the 20th century (E = mc²) informs barcode creation.
Send your comments to hspecht@parallelsq.org. Feedback and thoughts are received with thanks!

